Examples of Freundlich Group Projects
Representative projects leverage the following techniques: machine learning, computational modeling and drug design, chemical synthesis, intrabacterial drug metabolism (IBDM), biochemistry, microbiology, and molecular biology.
A. Novel chemical probes of bacterial biology
I. Initiated by our machine learning platform
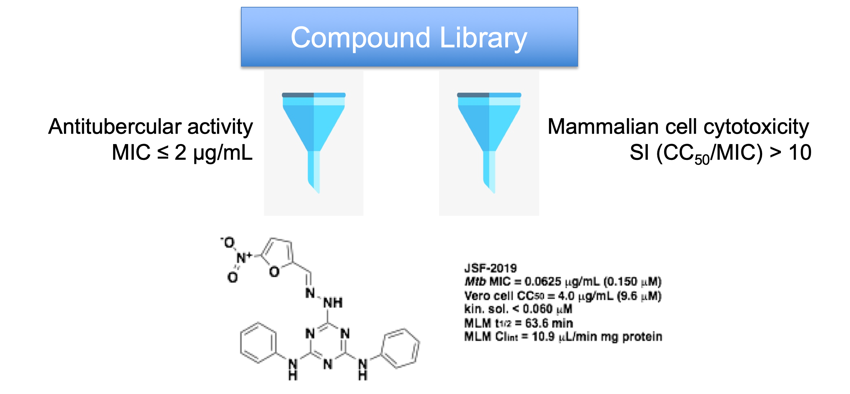 We assert that the research community can more optimally leverage the large data sets created over the last century to explore small molecule inhibition of the growth of bacteria of global health relevance such as M. tuberculosis. In vitro and in vivo data may be utilized to train machine learning models. These models enable exploration of new chemical space more efficiently than empirical high-throughput screening. We believe that such molecules, given their novel chemotypes amongst antibacterials, have a heightened probability of modulating novel biological targets. Thus, computational methods are being developed and then implemented to uncover new chemical probes of bacterial biology and drug discovery hits/leads.
We assert that the research community can more optimally leverage the large data sets created over the last century to explore small molecule inhibition of the growth of bacteria of global health relevance such as M. tuberculosis. In vitro and in vivo data may be utilized to train machine learning models. These models enable exploration of new chemical space more efficiently than empirical high-throughput screening. We believe that such molecules, given their novel chemotypes amongst antibacterials, have a heightened probability of modulating novel biological targets. Thus, computational methods are being developed and then implemented to uncover new chemical probes of bacterial biology and drug discovery hits/leads.
II. Initiated by other screening platforms
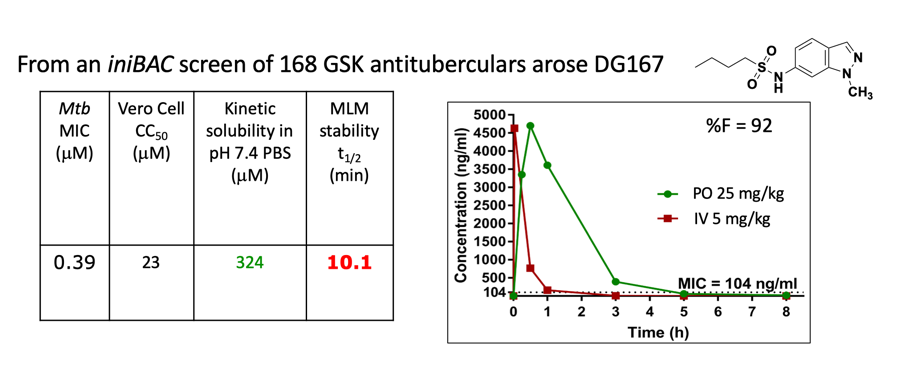 In collaboration we are exploring screening hits that exhibit pathway-specific inhibition within M. tuberculosis. Once hits are validated and profiled, we engage in mechanistic and chemical studies to determine molecular target. The products of this program have been novel antitubercular drug targets and small molecule chemical tools and drug discovery entities that potently modulate their activity.
In collaboration we are exploring screening hits that exhibit pathway-specific inhibition within M. tuberculosis. Once hits are validated and profiled, we engage in mechanistic and chemical studies to determine molecular target. The products of this program have been novel antitubercular drug targets and small molecule chemical tools and drug discovery entities that potently modulate their activity.
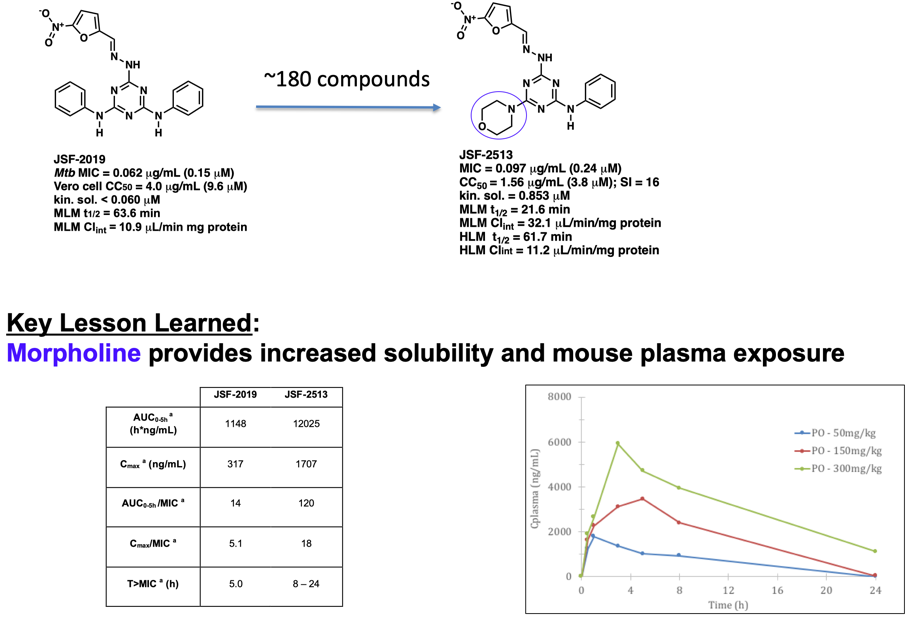 For both I. and II., the resulting novel antibacterials are optimized with respect to their efficacy, in vitro Absorption-Distribution-Metabolism-Excretion, in vivo pharmacokinetic, and toxicity profiles to obtain high-value chemical probes and drug discovery hits/leads. Here we can apply our machine learning platform to develop models for mouse liver microsome metabolism, aqueous solubility, Vero cell cytotoxicity, and other key properties. These models have afforded us alternative and complementary approaches (to traditional medicinal chemistry heuristics) to evolve hit compounds to afford valuable chemical tools and drug discovery leads in a time- and cost-efficient manner.
For both I. and II., the resulting novel antibacterials are optimized with respect to their efficacy, in vitro Absorption-Distribution-Metabolism-Excretion, in vivo pharmacokinetic, and toxicity profiles to obtain high-value chemical probes and drug discovery hits/leads. Here we can apply our machine learning platform to develop models for mouse liver microsome metabolism, aqueous solubility, Vero cell cytotoxicity, and other key properties. These models have afforded us alternative and complementary approaches (to traditional medicinal chemistry heuristics) to evolve hit compounds to afford valuable chemical tools and drug discovery leads in a time- and cost-efficient manner.
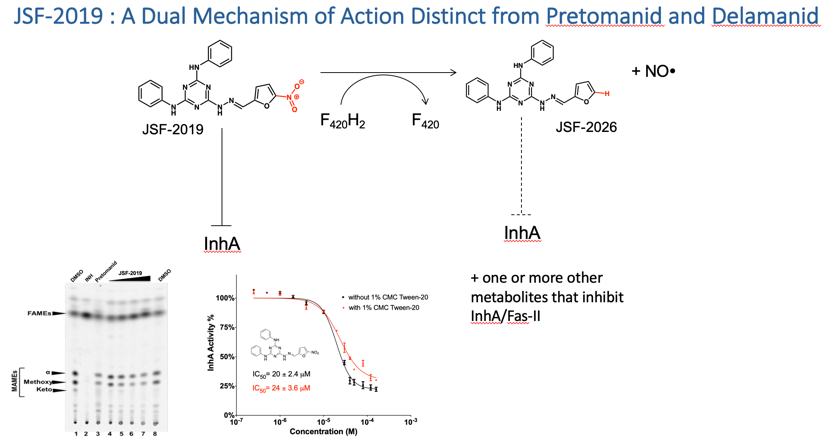 For both I. and II., mechanism of action is pursued through a combination of genetic, biochemical, and microbiology tools including our IBDM platform (Please see B.). The end result is a designation of molecular target/s for a given antibacterial and if necessary validation of the essentiality of that target (genetic and/or pharmacologic).
For both I. and II., mechanism of action is pursued through a combination of genetic, biochemical, and microbiology tools including our IBDM platform (Please see B.). The end result is a designation of molecular target/s for a given antibacterial and if necessary validation of the essentiality of that target (genetic and/or pharmacologic).
Representative Papers: Cell Chem Biol 2019, mBio 2018, Antimicrob Agents Chemother 2018, Pharm Res 2018, ACS Med Chem Lett 2017, Nat Chem Biol 2017, mBio 2017, Pharm Res 2016, Chem Biol 2013.
B. Exploration of the intrabacterial drug metabolism (IBDM) of drugs and chemical tools
Inspired by efforts in the metabolomics arena (in particular by the labs of Rabinowitz, Botstein, and Rhee) and driven to further probe the many antibacterial agents arising in the lab, we established the intrabacterial drug metabolism (IBDM) platform. To date, it has been instrumental in understanding the F420-dependent biotransformations of JSF-2019 and JSF-2164 and efforts are ongoing to apply this approach to other antibacterial agents. We anticipate this approach will enable the further evolution of these antibacterials while also enlightening us as to the intrabacterial biological pathways key to their metabolism.
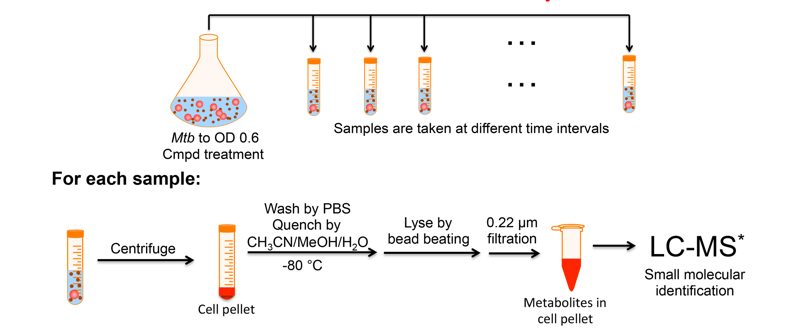
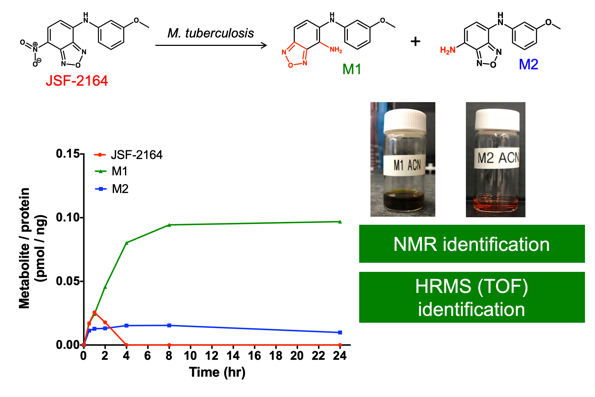
Recent Representative Papers: Cell Chem Biol 2019, ACS Infect Dis 2019, Trends Microbiol 2017.
