Human iPS Cell Core
The hiPS Cell Core, directed by Dr. Diego Fraidenraich, is a new initiative of the Department of Cell Biology and Molecular Medicine (CBMM), Rutgers-NJMS. The hiPS Cell Core facility, operating on a fee-for-service basis, is primarily a resource for CBMM investigators and investigators within the Rutgers community. The hiPS Cell Core provides GFP+ cardiomyocytes (CMs) derived from hiPS cells (hiPSC-CMs) (see details in links to the figures and movies).
Investigators are welcome to discuss scientific goals of the research involving the hiPSC-CMs with Dr. Fraidenraich, for further insight or collaborative/grant effort.
Our production approach utilizes an adapted method of a published protocol (GiWi method) that involves induction of differentiation using sequential inhibition of the GSK and WNT pathways, in RPMI medium without insulin (for details, please see:
Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, and Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013 January; 8(1): 162–175).
More information is forthcoming.
Acknowledgements
Dr. Radek Dobrowolski, Life Science Center, Rutgers University - Newark, provided us invaluable help with his much appreciated know-how and generous advice. Dr. Dobrowolski provided us with hiPS cells reprogrammed in his lab from dermal fibroblasts, which helped to establish the facility (http://www.ncas.rutgers.edu/radek-dobrowolski).
Dr. Jay Zhang, Department of Cardiology, University of Minnesota, Dr. Young-sup Yoon, Department of Cardiology, Emory University School of Medicine, Georgia and Dr. Mark Tomishima, SKI Stem Cell Research Facility, Memorial Sloan-Kettering Cancer Center, New York, were instrumental to our lab development through their kind hospitality and training in their labs. Dr. Jay Zhang’s gift of the hciPS cells reprogrammed from cardiac fibroblasts helped us to optimize the cardiomyocyte differentiation protocol in our lab.
We would also like to thank Dr. Nicole Dubois, Department of Developmental and Regenerative Biology, Icahn School of Medicine, Mount Sinai, New York for visiting our facility and giving us much needed technical advices and Dr. Jennifer Moore, RUCDR, Rutgers University, Piscataway for providing training in the state-of-the-art hiPSC reprogramming technique from human fibroblasts and lymphoblasts.
This new initiative could not have been possible without the support of Dr. Jun Sadoshima, from the Department of Cell Biology and Molecular Medicine at Rutgers – New Jersey Medical School.
Please inquire Dr. Diego Fraidenraich (Director) at fraidedi@njms.rutgers.edu or Dr. Erdene Baljinnyam (Manager) at baljiner@njms.rutgers.edu.
The order form should be downloaded, filled out, and submitted to:
Dr. Diego Fraidenraich/Dr. Erdene Baljinnyam
hiPS Cell Core Facility, MSB G-626
Department of Cell Biology and Molecular Medicine
NJMS, Rutgers University
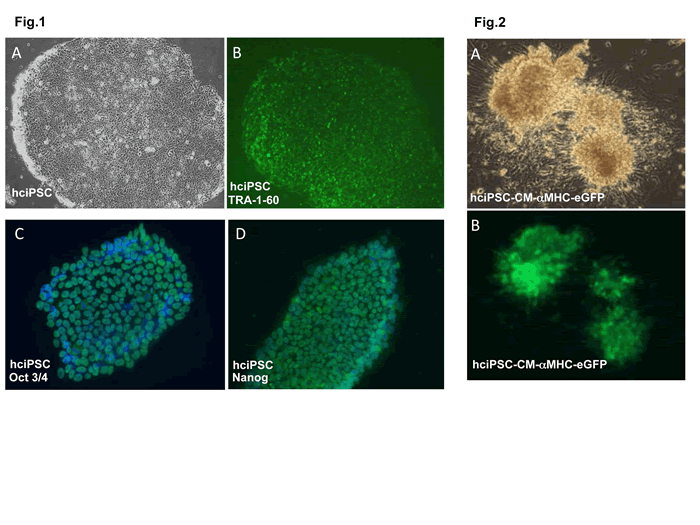
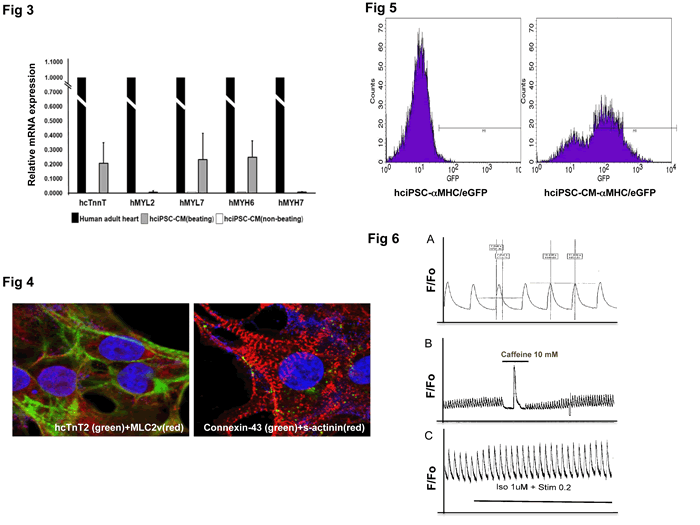
Fig 1. TRA-1-60 live staining for pluripotency in human cardiac fibroblast-derived induced pluripotent stem cells (hciPSCs). hciPSC were a kind gift of Dr. J. Zhang, University of Minnesota Medical School, Department of Medicine/Cardiology. hciPSC, A bright field, B TRA-1-60;
C. Pluripotency marker oct3/4 immunofluorescent (IMF) staining in hciPSCs; D. Pluripotency marker nanog IMF staining in hciPSCs.
Fig 2. hciPSC-CM-aMHC/eGFP+ cells. A. bright field, B. GFP.
Movie: synchronous spontaneous beating of hciPSC-CMs.
Fig 3. QRT-PCR analysis of hiPSC-CM contractile markers. Relative mRNA expression in beating vs non-beating hciPSC-CMs. Results are compared to those of human adult heart (arbitrary value = 1). Non-contractile markers THY1, hNEUROD, hPECAM were not expressed (not shown).
Fig 4. IMF staining of contractile markers (cTnT2, MLC2v and S-actinin) and Connexin-43 in hciPSC-CMs showing sarcomeric organization. Confocal images by courtesy of Dr. Luke Fritzky, Rutgers-NJMS, Cancer Center, Confocal Imaging Facility.
Fig 5. GFP-based FACS analysis of MHC/GFP-hciPSC-CMs (right). Note that MHC does not drive GFP expression in undifferentiated hiPSCs (left). FACS analysis was performed at NJMS Flow Cytometry And Immunology Core Laboratory, Dr. Patricia Fitzgerald-Bocarsly, Dr. Sukhwinder Singh.
Fig 6. Calcium transient measurement in hciPSC-CMs, using fura-2 as fluorescent dye. Calcium experiments were done in the laboratory of Dr. Lai-Hua Xie, NJMS Cell Biology and Molecular Medicine. A. Calcium transient in spontaneously beating hci-PSC-CM, T50=300 ms; B. Calcium release from SR upon caffeine 10 mM stimulation; C. Calcium transient response to Isoproterenol and pacing.
Click here for PDF of above figures
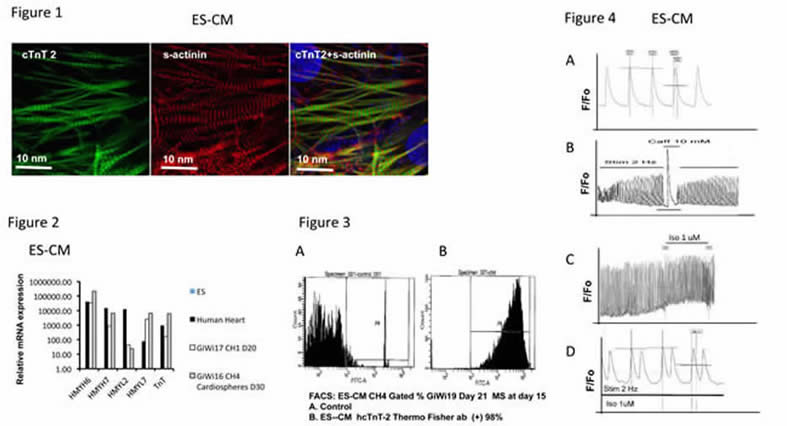
Fig 1. IMF staining of contractile markers cTnT2 (left) and S-actinin (center), and their merged image (right) in ES-CM showing sarcomeric organization.
Fig 2. QRT-PCR analysis of ES-CM contractile markers. Relative mRNA expression in beating monolayer vs cardiosphere ES-CMs. Results are compared to those of ESC (arbitrary value = 1). Non-contractile markers THY1, hNEUROD, hPECAM were not expressed (not shown).
Fig 3. FACS analysis of ES-CMs using an antibody reactive with hcTnT-2. ES-CM after metabolic selection (MS) at day 15. A. Control B. ES-CM hcTnT-2 (+) cells comprise 98% of the total cell number.
Fig 4. Calcium transient measurement in ES-CMs. A. Spontaneous calcium transients in ES-CM ; B. Calcium release from SR upon caffeine stimulation; C. Calcium transient response to Isoproterenol shows increased frequency. D. Calcium transient response to Isoproterenol shows ectopic peaks.
